A team of scientists from Italy and the USA has recently explored the maturation pattern of conventional dendritic cells (cDCs) during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Their findings reveal that the virus efficiently escapes host immune responses by directly interacting with cDCs and disrupting T cell activation. The study is currently available on the bioRxiv* preprint server.
Background
Since its emergence in December 2019 in China, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has infected 114 million people and claimed 2.5 million lives worldwide. In its milder form, COVID-19 is mainly associated with fever, dry cough, sore throat etc. However, a severe form of the disease often leads to serious health complications characterized by cardiovascular, pulmonary, and metabolic disorders. In general, COVID-19 patients with severe symptoms exhibit severe immune impairment characterized by aberrant production of proinflammatory mediators and alteration in dendritic cell populations in the peripheral blood.
In the context of any microbial infection, dendritic cells (DCs) are particularly important because of their significant involvement in T cell activation and antigen presentation. These cells directly sense pathogens and induce the production of proinflammatory mediators necessary for the initiation of adaptive immune responses. There are two subtypes of conventional DCs, namely cDC1 and cDC2. The cDC2 subtype is further divided into two subsets, namely DC2 and DC3. While cDC1s are specialized for antigen cross-presentation, cDC2s can induce a wide-range of immune responses by expressing different types of pattern recognition receptors (PRRs). There is evidence suggesting that SARS-CoV-2 infection causes a reduction in both the number and functionality of cDCs in the blood.
In the current study, scientists have explored the impact of SARS-CoV-2 infection on the maturation pattern of different cDC subtypes. They have specifically analyzed the peripheral blood cDCs isolated from mild and severe COVID-19 patients and healthy individuals.
Important observations
The scientists observed a reduction in cDC subtypes (cDC1s, DC2s, and DC3s) in the peripheral blood of COVID-19 patients compared to that in healthy individuals. Interestingly, the findings revealed that although the total population of DC3s showed a decreasing trend, an increasing trend in the proportion of inflammatory DC3s was observed in COVID-19 patients.
To determine the transcriptional response of different cDC subtypes to SARS-CoV-2 infection, the scientists thoroughly analyzed three different single-cell transcriptomics datasets. Their analysis revealed an upregulation of interferon-stimulated genes in all cDC subtypes isolated from COVID-19 patients.
Furthermore, they conducted a Principal Component Analysis using the single-cell dataset for an in-depth characterization of interferon-stimulated genes and inflammatory molecule-encoding genes in cDC subtypes. Their findings revealed significantly increased and decreased expressions of interferon-stimulated genes in cDCs from severe and mild COVID-19 patients, respectively. In contrast, a progressive reduction in the expressions of inflammatory molecule-encoding genes was observed with disease severity. These observations indicate an inverse correlation between the expression patterns of interferon-stimulated genes and inflammatory molecule-encoding genes. With further analysis, the scientists observed that type I interferon response stimulated by SARS-CoV-2 infection could equally affect the DC2 and DC3 subsets.
Besides interferon signaling pathways, an induction in PI3K-AKT-mTOR signaling pathway and a reduction in expressions of genes encoding major histocompatibility complex (MHC) class II molecules were observed in COVID-19 patients.
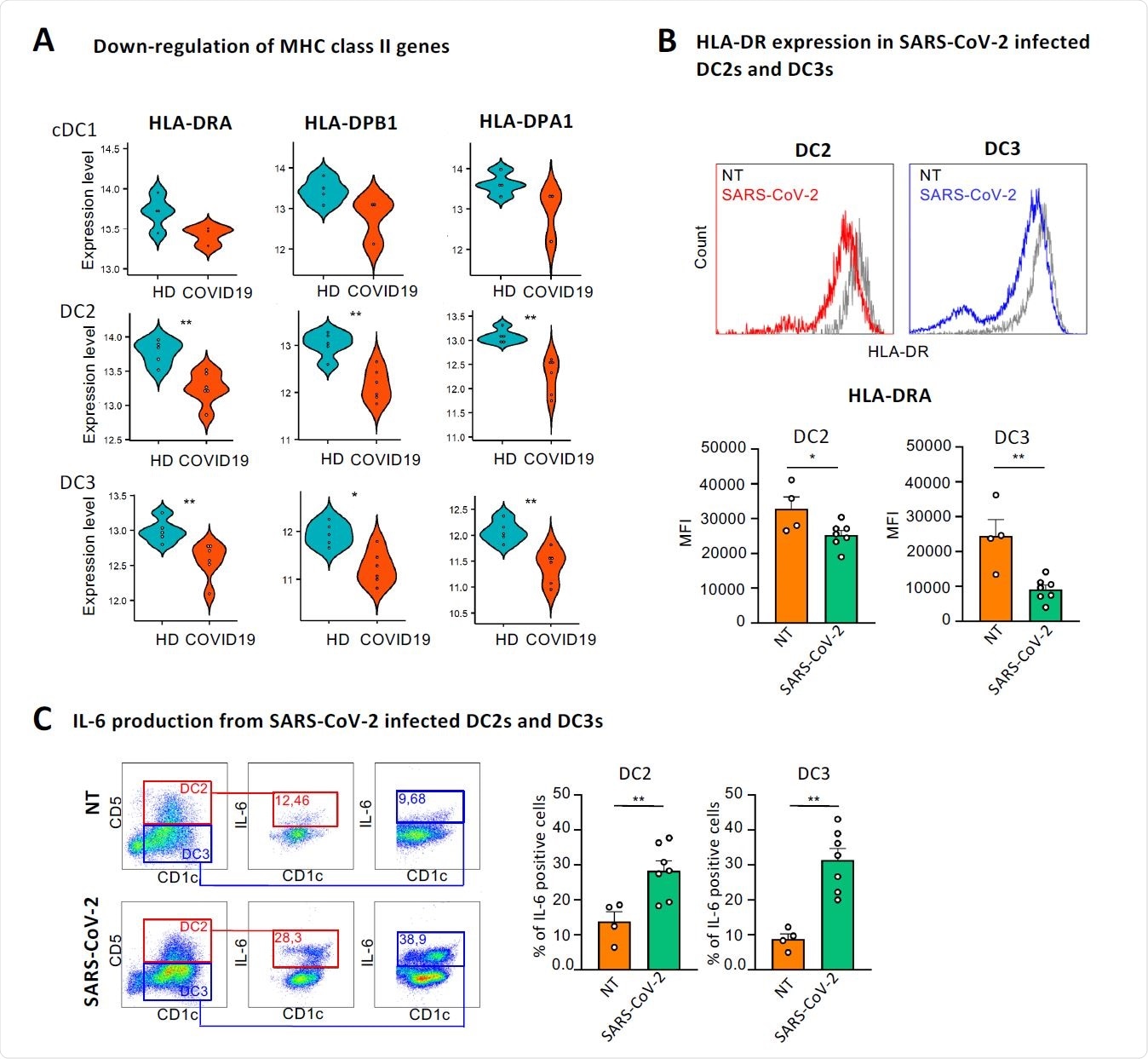
To explore the mechanistic details of SARS-CoV-2-induced alteration in immune signatures of cDCs, they isolated cDC2s from healthy individuals and assessed their response to the virus. The findings revealed that SARS-CoV-2 significantly reduced the expression of MHC class II molecules and increased the expression of IL-6 in both DC2 and DC3 subsets.
This indicates that the virus directly modulates the maturation process of cDCs to escape host immune responses. With further analysis, the scientists identified a set of 52 genes that were differentially expressed in DC3s compared to DC2s in response to SARS-CoV-2. By conducting a separate set of experiments using transcriptomics data of patients with bacterial sepsis, they identified a set of 203 genes that were differentially expressed in DC3s in response to bacterial infection.
These observations indicate that the prominent differences between DC2s and DC3s observed during bacterial infection are highly attenuated during SARS-CoV-2 infection. The similarity in responses of DC2s and DC3s in COVID-19 patients further indicates that a direct viral interaction rather than an exposure to infection-induced inflammatory mediators is responsible for the observed maturation patterns of these cells. Because of the variation in expressions of receptors like CD14 in DC2s and DC3, these two subsets could have responded differentially in response to exposure to inflammatory mediators.
Study significance
Overall, the study findings reveal that SARS-CoV-2 directly interacts with conventional DCs and suppresses the signaling cascade necessary for T cell activation. Such modifications at the genetic and molecular level subsequently facilitate the virus to evade host immune responses.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Marongiu L. 2021. Maturation signatures of conventional dendritic cells in COVID-19 reflect direct viral sensing. BioRxiv. doi: https://doi.org/10.1101/2021.03.03.433597, https://www.biorxiv.org/content/10.1101/2021.03.03.433597v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: Antigen, Blood, CD3, Cell, Coronavirus, Coronavirus Disease COVID-19, Cough, Dendritic Cell, Fever, Genes, Genetic, Metabolic Disorders, Molecule, Pathogen, Quantitative Analysis, Respiratory, SARS, SARS-CoV-2, Sepsis, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Signaling Pathway, Sore Throat, Syndrome, Throat, Transcriptomics, Virus

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article
