One in nine women in the developed world will be diagnosed with breast cancer at some point in her life. The prevalence of breast cancer is increasing, an effect caused in part by the modern lifestyle and increased lifespans. Thankfully, treatments are becoming more efficient and more personalized. However, what isn’t increasing—and is in fact decreasing—is the number of pathologists, or the doctors whose specialization is examining body tissues to provide the specific diagnosis necessary for personalized medicine.
A team of researchers at the Technion-Israel Institute of Technology has therefore made it their quest to turn computers into effective pathologists’ assistants, simplifying and improving the human doctors’ work. Their new study was recently published in Nature Communications.
The specific task that Dr. Gil Shamai and Amir Livne from the lab of Professor Ron Kimmel from the Henry and Marilyn Taub Faculty of Computer Science at the Technion set out to achieve lies within the realm of immunotherapy. Immunotherapy has been gaining prominence in recent years as an effective, sometimes even game-changing treatment for several types of cancer. The basis of this form of therapy is encouraging the body’s own immune system to attack the tumor. However, such therapy must be personalized, as the correct medication must be administered to the patients who stand to benefit from it based on the specific characteristics of the tumor.
Multiple natural mechanisms prevent our immune systems from attacking our own bodies. These mechanisms are often exploited by cancer tumors to evade the immune system. One such mechanism is related to the PD-L1 protein—some tumors display it, and it acts as a sort of password by erroneously convincing the immune system that the cancer should not be attacked. Specific immunotherapy for PD-L1 can persuade the immune system to ignore this particular password, but of course would only be effective when the tumor expresses the PD-L1.
It is a pathologist’s task to determine whether a patient’s tumor expresses PD-L1. Expensive chemical markers are used to stain a biopsy taken from the tumor in order to obtain the answer. The process is non-trivial, time-consuming, and at times inconsistent. Dr. Shamai and his team took a different approach. In recent years, it has become an FDA-approved practice for biopsies to be scanned so they can be used for digital pathological analysis. Amir Livne, Dr. Shamai and Prof. Kimmel decided to see if a neural network could use these scans to make the diagnosis without requiring additional processes. “They told us it couldn’t be done,” the team said, “so of course, we had to prove them wrong.”
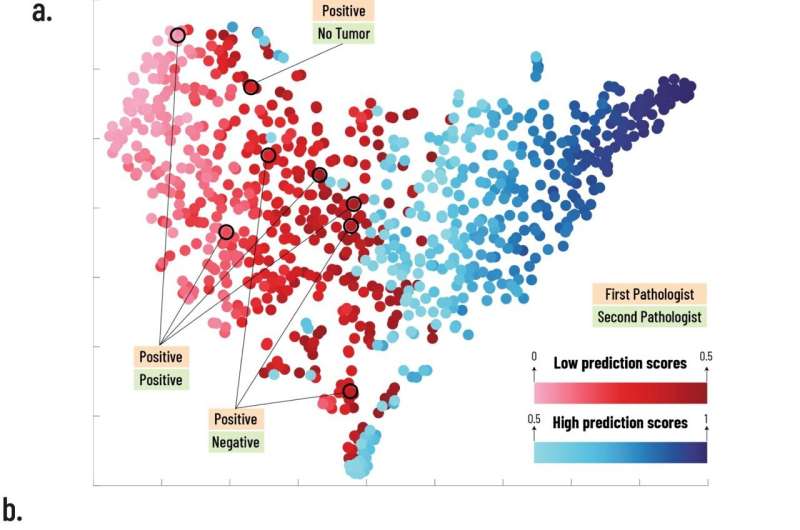
Neural networks are trained in a manner similar to how children learn: they are presented with multiple tagged examples. A child is shown many dogs and various other things, and from these examples forms an idea of what “dog” is. The neural network Prof. Kimmel’s team developed was presented with digital biopsy images from 3,376 patients that were tagged as either expressing or not expressing PD-L1. After preliminary validation, it was asked to determine whether additional clinical trial biopsy images from 275 patients were positive or negative for PD-L1. It performed better than expected: For 70% of the patients, it was able to confidently and correctly determine the answer. For the remaining 30% of the patients, the program could not find the visual patterns that would enable it to decide one way or the other. Interestingly, in the cases where the artificial intelligence (AI) disagreed with the human pathologist’s determination, a second test proved the AI to be right.
“This is a momentous achievement,” Prof. Kimmel explained. “The variations that the computer found—they are not distinguishable to the human eye. Cells arrange themselves differently if they present PD-L1 or not, but the differences are so small that even a trained pathologist can’t confidently identify them. Now our neural network can.”
This achievement is the work of a team comprised of Dr. Gil Shamai and graduate student Amir Livne, who developed the technology and designed the experiments, Dr. António Polónia from the Institute of Molecular Pathology and Immunology of the University of Porto, Portugal, Professor Edmond Sabo and Dr. Alexandra Cretu from Carmel Medical Center in Haifa, Israel, who are expert pathologists that conducted the research, and with the support of Professor Gil Bar-Sela, head of oncology and hematology division at Haemek Medical Center in Afula, Israel.
“It’s an amazing opportunity to bring together artificial intelligence and medicine,” Dr. Shamai said. “I love mathematics, I love developing algorithms. Being able to use my skills to help people, to advance medicine—it’s more than I expected when I started out as a computer science student.” He is now leading a team of 15 researchers, who are taking this project to the next level.
“We expect AI to become a powerful tool in doctors’ hands,” shared Prof. Kimmel. “AI can assist in making or verifying a diagnosis, it can help match the treatment to the individual patient, it can offer a prognosis. I do not think it can—or should—replace the human doctor. But it can make some elements of doctors’ work simpler, faster, and more precise.”
More information:
Gil Shamai et al, Deep learning-based image analysis predicts PD-L1 status from H&E-stained histopathology images in breast cancer, Nature Communications (2022). DOI: 10.1038/s41467-022-34275-9
Journal information:
Nature Communications
Source: Read Full Article
