The coronavirus disease 2019 (COVID-19) can significantly alter host immune and inflammation function. In fact, many researchers have suggested that some of the symptoms observed in ‘long COVID could be related to this phenomenon. In particular, alterations in the myeloid immune compartment have been frequently reported in COVID-19; however, these effects are rarely investigated.
In a recent study published on the preprint server bioRxiv*, researchers investigate classical CD14+ monocytes in healthy and infected individuals to better understand how infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can cause dysfunction in these immune cells.
Study: Transcriptional reprogramming from innate immune functions to a pro-thrombotic signature upon SARS-CoV-2 sensing by monocytes in COVID-19. Image Credit: Kateryna Kon / Shutterstock.com
Study findings
To characterize the ex vivo phenotype of CD14+ monocytes in both uninfected healthy individuals and patients presenting with mild to moderate COVID-19, several different markers were examined through high dimensional flow cytometry. Dimensionality reduction tools were used to demonstrate that while there was some overlap in the global phenotype between the different groups, monocytes from healthy individuals were significantly distinct from monocytes isolated from individuals suffering from mild and moderate COVID-19.
Moderate COVID-19 monocytes could be identified by decreased levels of expression of human leukocyte antigen DR phenotype (HLA-DR), as well as increased expression of HLA-ABC as compared to the other two groups. These monocytes also exhibit increased expression of CD301. Other evidence of an altered activation profile was observed through the reduced expression of CD86 and increased expression of T-cell immunoglobulin (TIM-3) and programmed cell death protein 1 (PD-I).
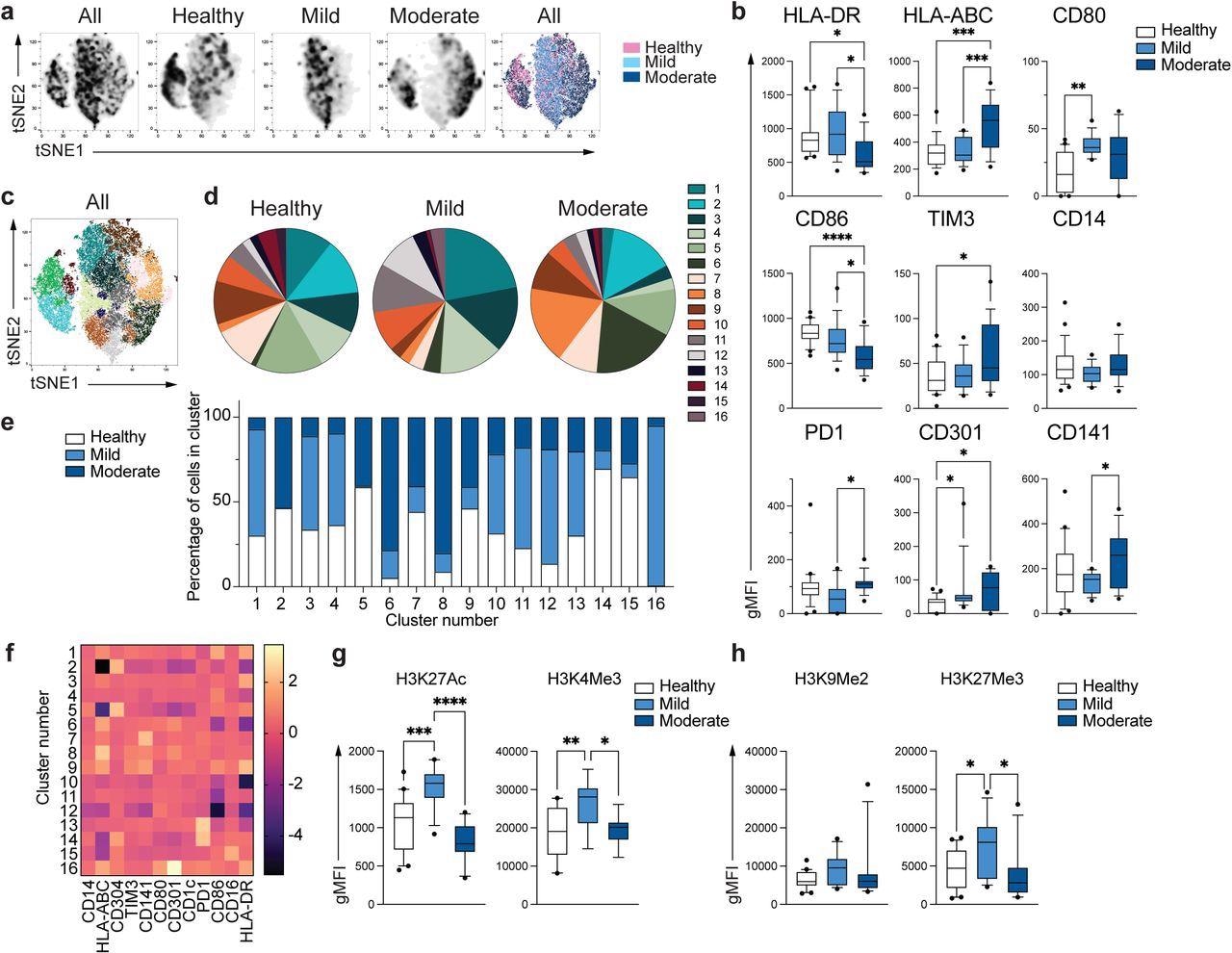
Unique phenotype of COVID-19 monocytes. a. tSNE plots obtained from a concatenated sample consisting of PBMC from n=15 healthy individuals, n=15 mild and n=15 moderate COVID-19 patients. b. Box and whiskers plots summarizing the median gMFI of the receptors analyzed. The box extends from the 25th to the 75th percentile and the whiskers are drawn down to the 10th percentile and up to the 90th percentile. Points below and above the whiskers are drawn as individual points (n=25 healthy, n=15 mild and n=17 moderate COVID-19 individuals). c. tSNE plots depicting the cell clusters identified by Phenograph from the concatenated sample in a. d. Pie charts show the fraction of cells within each identified cell cluster in each patient group. e. Bars graph show the distribution (percentage) of cells from each patient group in each identified cell cluster. f. Heatmap of the expression of receptors per cell cluster displayed as modified z-scores using median values. g and h. Summary of expression of activating (g) and repressive (h) histone marks in monocytes from healthy individuals (n=20), mild (n=15) and moderate (n=11) COVID-19 patients. One-way ANOVA with Tukey’s correction for multiple comparisons for b, g, h. *P<0.05, **p<0.005, ***p<0.001, ****p<0.0001.
Clustering algorithms using 12 phenotypic markers that had previously been examined were used to define the phenotypic differences between healthy individuals and COVID-19 patients. To this end, 16 subpopulations of monocytes with distinctive differences in distributions between healthy and infected patients were identified.
The expansion of specific monocyte subpopulations was also found to be different between mild and moderate patients. When the expression levels of the markers that defined each cluster were normalized, the phenotype of clusters 6 and 8, which contained predominantly moderate monocytes, were driven by the downregulation of CD86 and HLA-DR or upregulation of HLA-ABC, respectively.
Principal component analysis (PCA) was then applied to examine the global distribution of gene expression profiles from monocytes from infected and healthy individuals. To this end, there were distinct differences between these groups, with genes encoding soluble factors, chemokines, and class II molecules as the main genes contributing to the separation between monocytes from different groups.
Differential gene analysis showed 422 upregulated and 187 downregulated genes in COVID-19 monocytes. Through the use of pathway enrichment analysis, glycolysis was found to be the most enriched pathway, followed by the metabolism of lipids and lipoproteins. Interferon (IFN)/cytokine signaling was also present.
Taking a closer look at the directionality of expression of these pathways revealed variable expression patterns between COVID-19 monocytes, including several type I IFN-stimulated genes and metabolic enzymes. The highest expressed IFN-related gene was IF127, which was considered to be a biomarker of early SARS-CoV-2 infection.
After discovering that the only significantly downregulated pathway in COVID-19 monocytes was glycolysis, SCENITH was used to profile the metabolism of monocytes from different groups. Puromycin incorporation, which was used as a marker of protein synthesis, was significantly decreased in moderate COVID-19 monocytes, thus suggesting decreased metabolic activity.
Several attempts to examine the capacity of monocytes to sense and respond to SARS-CoV-2 ex vivo were also made. To this end, the stimulation of monocytes from healthy individuals using SARS-CoV-2 revealed a significant increase in both tumor necrosis factor (TNF) and interleukin 10 (IL-10) individuals.
However, in COVID-19 monocytes, significantly less TNF was produced. This reaction was not SARS-CoV-2 specific, as stimulation with common coronaviruses and bacterial lipopolysaccharide also led to significantly reduced TNF production. COVID-19 monocytes also failed to increase CD86 expression after stimulation.
Ribonucleic acid sequencing (RNA-Seq) on stimulated monocytes from both groups of individuals, followed by PCA, separated COVID-19 monocytes from healthy monocytes, with 1,437 upregulated and 2,073 downregulated genes in activated COVID-19 monocytes. Pathway enrichment confirmed previous findings, with type I IFN signaling, cytokines signaling, and interactions between lymphoid and non-lymphoid cells all differentially expressed.
Conclusions
The current study reveals that distinct populations of circulating monocytes are significantly enriched in mild and moderate COVID-19 patients. Furthermore, a more profound dysfunction in moderate as compared to mild COVID-19 monocytes was also observed.
Thus, the researchers confirm dysfunction in the metabolism in COVID-19 monocytes and suggest that this is partly responsible for the reduced responsiveness of these monocytes to pathogens. Taken together, the factors identified in this study could help inform healthcare options and potentially lead to future treatment targets for COVID-19.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Maher, A. K., Burnham, K. L., Jones, E., et al. (2022). Transcriptional reprogramming from innate immune functions to a pro-thrombotic signature upon SARS-CoV-2 sensing by monocytes in COVID-19. bioRxiv. doi:10.1101/2022.04.03.486830. https://www.biorxiv.org/content/10.1101/2022.04.03.486830v1.full.
Posted in: Molecular & Structural Biology | Cell Biology | Medical Science News | Medical Research News | Disease/Infection News
Tags: Antigen, Biomarker, Cell, Cell Death, Chemokines, Coronavirus, Coronavirus Disease COVID-19, Cytokine, Cytokines, Cytometry, Ex Vivo, Flow Cytometry, Gene, Gene Expression, Genes, Glycolysis, Healthcare, Human Leukocyte Antigen, Immunoglobulin, Inflammation, Interferon, Interleukin, Leukocyte, Lipids, Metabolism, Monocyte, Necrosis, Phenotype, Programmed Cell Death, Protein, Protein Synthesis, Respiratory, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, T-Cell, Tumor, Tumor Necrosis Factor

Written by
Sam Hancock
Sam completed his MSci in Genetics at the University of Nottingham in 2019, fuelled initially by an interest in genetic ageing. As part of his degree, he also investigated the role of rnh genes in originless replication in archaea.
Source: Read Full Article