A newly identified bacteria and a common virus may be the underlying cause of infection-induced hydrocephalus in Uganda, according to an international team of researchers.
“Thirteen years ago, while visiting Uganda and seeing a stream of kids with hydrocephalus after infection I asked the doctors, ‘What is the biggest problem you have that you can’t solve?'” said Steven J. Schiff, Brush Chair Professor of Engineering and professor of engineering science and mechanics, neurosurgery and physics, Penn State. “‘Why don’t you figure out what makes these kids sick?’ was the reply.”
By that time, the doctors at CURE Children’s Hospital of Uganda, had seen more than a thousand infants with infection-caused hydrocephalus and were unable to culture a single thing in the laboratory. They have now seen over 8,000 similar children in this one, small Ugandan hospital.
“Hydrocephalus is the most common childhood neurosurgical condition that we see in the population that we serve,” said Edith Mbabazi-Kabachelor, director of research, CURE Children’s Hospital of Uganda. “If hydrocephalus is left untreated in children less than two years old, the progressive increase in head size will lead to further brain damage, resulting in the majority of these children dying, and those that survive being left with severe cognitive and physical disability.”
Severe systemic bacterial infection during the first four weeks of life accounts for an estimated 680,000 to 750,000 yearly neonatal deaths worldwide. Hydrocephalus is the most common brain disorder in childhood and the largest single cause of childhood hydrocephalus is neonatal infection, accounting for an estimated 160,000 yearly cases, said Schiff.
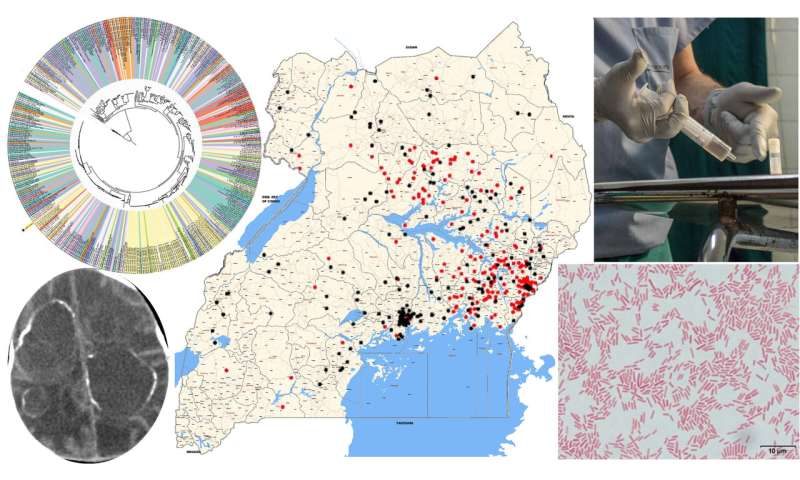
Over a 5-year study in Uganda, supported by the U.S. National Institutes of Health, using advanced genomic techniques the team uncovered the major bacterial and viral underpinnings of these infections, the researchers report today (Sept. 30) in Science Translational Medicine.
Schiff and his team have studied this problem for more than 10 years, but in the last five years, they took a different approach, using DNA and RNA sequencing techniques to identify the causative agents. The researchers looked at blood and cerebrospinal fluid drawn from 100 cases of post-infectious hydrocephalus and control patients without infection in Uganda. There were 64 infants with post-infectious hydrocephalus and 36 with non-post-infectious hydrocephalus. All infants were under three months old. The researchers prepared the samples in two ways—fresh-frozen and preserved—and they sent samples to two different laboratories in the U.S., where samples were analyzed with different techniques. This was to ensure valid and reproducible results.
“We found this weird bacteria dominating,” said Schiff.
The bacteria was a previously unidentified strain of Paenibacillus thiaminolyticus, now named Mbale after the city where the CURE Children’s Hospital is located.
“The initial link between hydrocephalus and Paenibacillus was made through high-throughput sequencing and PCR analyses at the Center for Infection and Immunity in the Mailman School of Public Health at Columbia University, a renowned center led by W. Ian Lipkin,” said Schiff.
High-throughput sequencing allows sequencing of more than one DNA molecule at the same time, and PCR analysis multiplies existing DNA samples so that they are easier to analyze and identify.
“You build a field of dreams—in this case a platform for pathogen discovery—and wait for the right partner and the right project,” said W. Ian Lipkin, John Snow Professor of Epidemiology and director, Center for Infection and Immunity, Mailman School of Public Health, and professor of pathology and neurology, College of Physicians & Surgeons, Columbia University. “Steven Schiff is a remarkable investigator and this is such a project. It stands out for impact amongst hundreds we’ve done over a period of more than 30 years. Our team is delighted to have had an opportunity to help implicate an agent and contribute to control of this devastating disorder.”
The researchers managed to grow the difficult-to-culture new bacterial strain at Penn State, and tested it on mice. While the common variants of Paenibacillus are harmless, the Mbale strain was lethal to the mice.
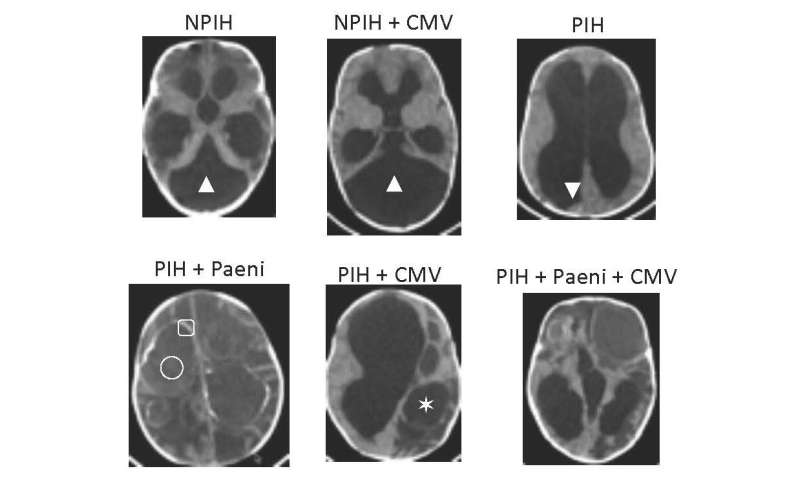
The researchers found the new bacterial strain in the cerebrospinal fluid of the infection-induced hydrocephalic children and then only in the youngest patients.
“While we tested infants up to three months old, we mostly identified the cause of infections in those less than six weeks of age,” said Schiff. “If we didn’t study them really early in life, then the infection had already burned out. Between 6 and 12 weeks there were very few positive results.”
Schiff was not satisfied with finding the proposed bacterial cause of the problem, he said. He reasoned that other diseases had both a bacterial and viral component and so the team looked for viral, fungal and parasitic genetic material. They found cytomegalovirus (CMV) in the cerebrospinal fluid of the infection-caused hydrocephalic infants, but not in that of the other hydrocephalus patients.
CMV is a common virus found around the world. The virus causes minor symptoms, if any, in most adults, although babies may be born with congenital CMV or acquire it early in life and be significantly harmed by neurological damage. The researchers only found CMV in the cerebrospinal fluid of babies with post-infection hydrocephalus.
While the researchers believe they have found the source of the infections that cause the high prevalence of hydrocephalus, they do not know where the babies encounter the new bacteria. According to Schiff, the bacteria may be soil- or water-born and more work is necessary to find the bacterial source.
The researchers are creating predictive models that, coupled with data they are now analyzing from thousands of infants and satellite-acquired rainfall to predict optimal treatment for individual locations. The researchers said they do not know if this particular bacterial virus combination exists outside this area of Uganda. However, the same strategy of using DNA and RNA to diagnose previously unknown causes of similar infections can be used in many other regions in the developing world where similar cases are seen.
Source: Read Full Article
