Since the onset of the coronavirus disease 2019 (COVID-19) pandemic, several variants of its etiological agent of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have emerged.
The emergence of these variants, particularly the most recent Omicron variant of concern (VOC), has raised concerns regarding their ability to escape from vaccine-induced antibody immunity. However, there remains limited information available as to whether T-cell immunity is also impacted by the Omicron variant.
Study: Ablation of CD8 + T-cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron. Image Credit: Giovanni Cancemi / Shutterstock.com
Most T-cell epitopes have been reported to be conserved, with the majority of vaccine recipients found to conserve T-cell immunity. However, several studies suggest that SARS-CoV-2 variants could escape the T-cell response induced in some convalescent individuals.
In a preprint* research paper under consideration for publication in the Nature Portfolio Journal, researchers describe the impact of changes in the Omicron variant on a uniquely immunodominant spike-encoded CD8+ T-cell epitope.
About the study
The study recruited participants who were over 18 years of age and provided informed consent to participate in the study. The participants were divided into two cohorts, which included those who had been clinically diagnosed with COVID-19 and those who had not been exposed to SARS-CoV-2 but had received two doses of vaccine.
Blood samples were collected from convalescent individuals after symptoms resolved, whereas samples were collected from unvaccinated individuals 28 days after they had received their second dose. Thereafter, human leukocyte antigen (HLA) typing was performed on each of the collected samples.
Expansion of SARS-CoV-2-specific T-cells was carried out with the help of pools of 15-mer peptides with an 11-amino-acid overlap. This was followed by intracellular cytokine assay, functional avidity assay, and HLA restriction assay.
Expression, refold, and purification of pHLA-A*29:02 complexes was carried out using pET-30a (+) DNA plasmids. A thermal stability assay was performed with the help of differential scanning fluorimetry. Finally, crystallization and structural determination of the HLA-A*29:02-peptide complex was carried out.
Study findings
Immunodominant CD8+ T cell responses to spike (S-1 pool), as well as ORF3A, were reported in all HLA-A*29:02 + participants. Both the S1 and ORF3A-specific CD8+ T-cell response was found to be higher in HLA-A*29:02 positive participants as compared to HLA-A*29:02 negative participants.
Spike-specific T-cell responses were found to target spike matrix pools that corresponded to overlapping peptides GFNCYFPLQSYGFQP and YFPLQSYGFQPTNGV. The HLA-A*29:02-restricted epitope, YFPLQSYGF, was also found to be responsible for the immunodominant response.
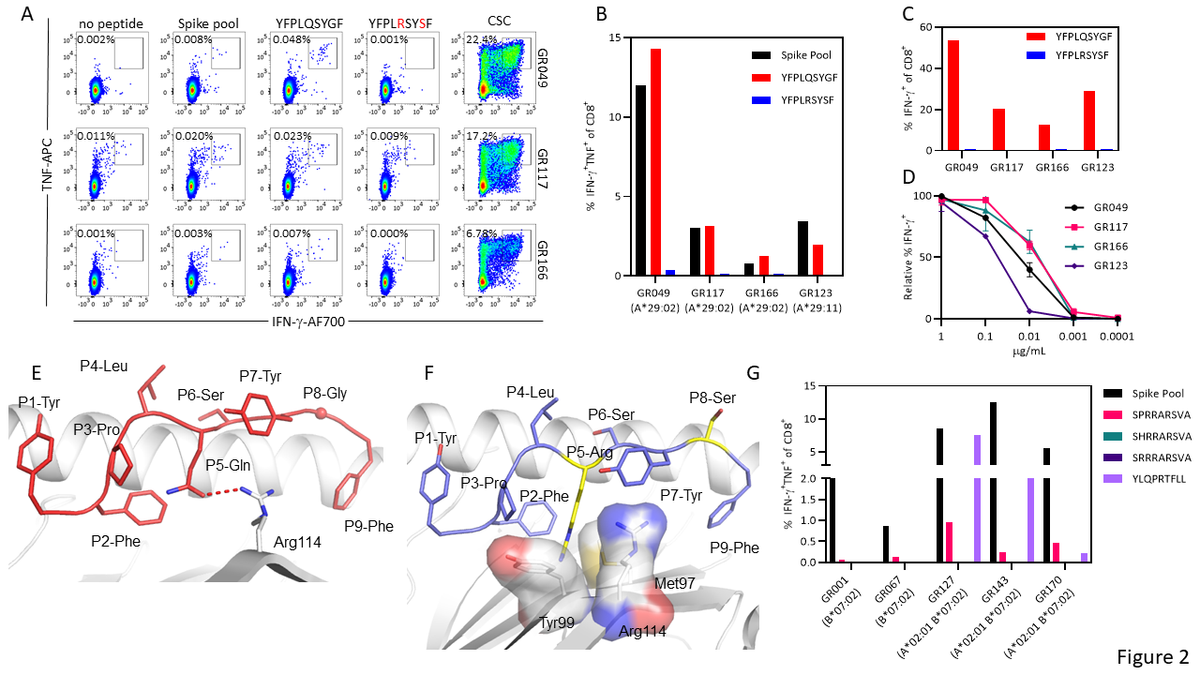
Recognition of the immunodominant HLA-A*29:02-restricted epitope in vaccinated participants. (A) Flow cytometry plots displaying the frequency of IFN-g-producing CD8+ T cells from PBMC of vaccinated HLAA*29:02+ individuals. (B) Frequency of IFN-g-producing CD8+ T cells following in vitro culture of PBMC from four vaccinated individuals (A*29:02 n=3; A*29:11 n=1) with spike overlapping peptide pools. (C) Frequency of IFN-g-producing CD8+ T cells following in vitro stimulation with either YFPLQSYGF or YFPLRSYSF and recall with cognate peptide. (D) Frequency of IFN-g-producing CD8+ T cells in response to serial dilutions of YFPLQSYGF. (E) Crystal structure of the HLA-A*29:02-YFPLRSYSF complex with the HLA heavy chain in white cartoon and the peptide in red cartoon and stick. The P8-Gly Ca atom is represented by a sphere, and the hydrogen bond by a red dashed line. (F) Model of the YFP variant peptide (blue cartoon and stick) based on the YFP peptide conformation showing the P5-Arg and P8-Ser substitutions in yellow stick. The HLA Met97, Tyr99, and Arg114 are represented as stick and transparent surface to show the steric clashes with the P5-Arg occurring if the variant adopted the same conformation as the cognate peptide. (G) Frequency of IFN-g-producing CD8+ T cells following in vitro culture of PBMC from five HLA-B*07:02 vaccinated individuals with spike overlapping peptide pools.
The ORF3a-specific T cell response was found to target matrix pools 7, 8, and 15 that corresponded to overlapping peptides VVLHSYFTSDYYQLY and SYFTSDYYQLYSTQL, which also contained the HLA-A*29:02-restricted epitope YFTSDYYQLY.
The ORF3A-encoded peptide was conserved, while amino acid substitutions at positions 5 and 8 were reported in the spike-encoded epitope for the Omicron variant. Substitution at position 8 did not appear to impact HLA-A*29:02, while a mutation in position 5 can change the peptide structure, as well as weaken its binding to HLA-A*29:02.
Low-frequency interferon-gamma (IFN-g)+ tumor necrosis factor (TNF)+ CD8+ T-cell responses against both the spike overlapping peptide pools and the YFPLQSYGF epitope were reported in three vaccinated individuals with HLA-A*29:02. However, CD8+ T-cells capable of recognizing the Omicron variant were absent in them.
Analysis of the HLA-B*07:02-restricted epitope SPRRARSVA in five HLA-B*07:02-positive vaccine recipients indicated that this epitope contained mutations mostly at position 2 and was detected in multiple variants. Although T-cells from vaccinated individuals could not recognize the variant peptide, other spike-specific T-cell responses were detected. This suggests that the T-cell immunodominance in HLA-distinct individuals was complex.
Conclusions
The current study demonstrates that HLA-A29-positive individuals were associated with skewed T-cell responses towards a single dominant epitope.
In most populations, the HLA-A29 frequencies are found to be mostly lower, while some populations in Africa have reported levels as high as 24%. Therefore, genetic variation in the newly emerging SARS-CoV-2 variants can lead to escape from cellular immunity, especially in the case of ethnic groups in which HLA alleles impacted by epitope mutations are dominant.
*Important notice
Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Smith, C., Swaminathan, S., Lineburg, K., et al. (2022). Ablation of CD8 + T-cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron. Nature Portfolio Journal. doi:10.21203/rs.3.rs-1289622/v1. https://www.researchsquare.com/article/rs-1289622/v1.
Posted in: Molecular & Structural Biology | Medical Science News | Medical Research News | Disease/Infection News
Tags: Amino Acid, Antibody, Antigen, Assay, Blood, Cell, Coronavirus, Coronavirus Disease COVID-19, covid-19, Cytokine, DNA, Frequency, Genetic, Human Leukocyte Antigen, immunity, Interferon, Interferon-gamma, Intracellular, Leukocyte, Mutation, Necrosis, Omicron, Pandemic, Peptides, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, T-Cell, Tumor, Tumor Necrosis Factor, Vaccine

Written by
Suchandrima Bhowmik
Suchandrima has a Bachelor of Science (B.Sc.) degree in Microbiology and a Master of Science (M.Sc.) degree in Microbiology from the University of Calcutta, India. The study of health and diseases was always very important to her. In addition to Microbiology, she also gained extensive knowledge in Biochemistry, Immunology, Medical Microbiology, Metabolism, and Biotechnology as part of her master's degree.
Source: Read Full Article