Ribosomes are highly complex, macromolecular structures that fulfil the vital role of protein synthesis in all living cells across species, from bacteria to eukaryotes.
 Desigua | Shutterstock
Desigua | Shutterstock
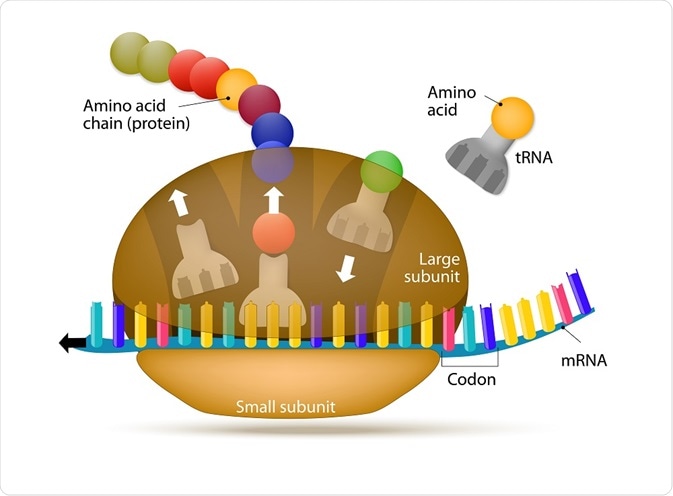
Ribosomes appear flattened and spherical in shape when viewed under an electron microscope, with a diameter ranging between 15 to 25 nm. These structures are comprised of two major ribonucleoprotein subunits. The smaller subunit mediates the correct interactions between three consecutive, in-frame base pairs, called codons and the complementary triplet called the anti-codon on the transfer RNA (tRNA).
The larger subunit, which is approximately twice the size of the smaller, contains the peptidyl-transferase center (PTC), which catalyzes the condensation reaction that mediates the formation of the peptide bond between amino acids in the growing polypeptide. The two subunits assemble to translate mRNA and disassemble when translation is complete.
Parts of the ribosome
The ribosomal structure is highly conserved between all species, but its exact composition depends on the organism. The yeast, Saccharomyces cerevisiae is the canonical model upon which the structural dynamics of the ribosome have been elucidated.
Yeast produce approximately 2000 per minute to satisfy the protein demand. Each ribosome is comprised of four ribosomal RNAs (rRNAs) and 79 ribosomal proteins (r‐proteins) necessary to produce a single functioning ribosome particle; an appreciable amount of energy is therefore necessary to ensure all ribosomes produced correctly.
Across all species, the ribosomal subunits are characterized by the Svedberg units, abbreviated S. These units are based on the rate at which subunits sediment in a solvent; concomitantly, this is derived from measurement of a parameter called the apparent sedimentation coefficient.
The simplest ribosome is from bacteria and is comprised of a 30S and 50S subunit which together form the 70S ribosome. Contrastingly, yeast ribosomes are comprised of a 40S and 60S subunit, which assemble to form the 80S structure.
Each subunit across species contains three sites that underpin the translation process. These are designated the A (aminoacyl), which accommodates the incoming tRNA which is primed with an amino acid (aminoacylated-tRNA); P (peptidyl), which holds the tRNA with the emerging, or nascent, peptide chain (peptidyl-tRNA); and E (exit), which mobilizes the emerging tRNA which has been stripped of its amino acid (deacylated-tRNA).
Relative to bacterial ribosomes, the eukaryotic ribosomes are large and more complex. They contain expansion segments (ES) as well as additional r-proteins and r-protein extensions. They contain over 5500 nucleotides of rRNA distributed in the small subunit (18S rRNA) and large subunit (5S, 5.8S, and 25S rRNA) alongside the 79 r-proteins (80 in other eukaryotes).
The full assignment and mapping of rRNAs and r-proteins were completed following the crystal structures obtained from both the small and large subunits Tetrahymena thermophila and the Saccharomyces cerevisiae 80S ribosome.
The RNA within ribosomes is organized by tertiary structural motifs, such as pseudoknots, and is responsible for carrying out the catalytic activity of the ribosome. The proteins surround the ribosome on the outside and are believed to stabilize its structure.
Free and membrane bound ribosomes
The location of protein synthesis depends on the ultimate purpose and destination of the protein. If the protein has a signal directing it to the endoplasmic reticulum (ER), then the ribosome will bind to the ER membrane. The same ribosome may disassemble and become a free ribosome when it begins translating its next protein. Free ribosomes synthesize proteins mainly for use within the cell. In eukaryotes, ribosomes are found predominantly attached to the ER.
Ribosomes bound to a membrane synthesize proteins for export outside the cell or for use in lysosomes. Membrane bound ribosomes are found in the ER. In fact, the presence of ribosomes gives the ER its signature “rough” appearance in electron micrographs.
Assembly
The assembly of the ribosome begins in the nucleolus, a region located in the nucleus. Here, both the genes for the r-proteins and rRNAs are translated concurrently. Once the r-proteins have been synthesized in the cytoplasm following translation, consequently associate with the pre-rRNA molecules in a co-transcriptional manner.
Owing to the enormous complexity of the ribosome, the assembly process employs upwards of 200 non-ribosomal biogenesis factors that function transiently. The first structure that results from pre-rRNA and r-protein association is called the 90S particle; following cleavage of the pre-rRNA, release of two particles, the pre‐60S and pre‐40S occurs.
These subunits subsequently acquire export competence as they undergo maturation in the nucleus. This maturation process continues in the cytoplasm after they pass across the nuclear envelope via the nuclear pore complexes (NPC). In the cytoplasm, the incorporation of more r-proteins licenses the ribosomal subunits to join and begin translation.
Whilst considerable insights into the structures of mature ribosomes have been made, less is known about the process of ribosomal assembly, particularly how stable expression, nuclear import, and assembly of r‐proteins occurs.
Sources
- British Society for Cell Biology-Ribosome
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (February 2011). "The structure of the eukaryotic ribosome at 3.0 Å resolution". Science. 334 (6062): 1524–1529.
- Rabl, Leibundgut, Ataide, Haag, Ban (February 2010). Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 331 (6018): 730–736. doi:10.1126/science.1198308. PMID 21205638
- Klinge, Voigts-Hoffmann, Leibundgut, Arpagaus, Ban (November 2011). "Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6". Science. 334 (6058): 941–948.
- Recht MI, Douthwaite S, Puglisi JD (1999). "Basis for bacterial specificity of action of aminoglycoside antibiotics". EMBO J. 18 (11): 3133–8. doi:10.1093/emboj/18.11.3133. PMC 1171394
- Ribosome assembly in eukaryotes.
- Molecular Biology of the Cell. 4th Ed., The Endoplasmic Reticulum.
Further Reading
- All Cellular Biology Content
- Structure and Function of the Cell Nucleus
- What Are Organelles?
- Protein Production: Initiation, Elongation and Termination
- Histone Modification
Last Updated: Apr 3, 2019

Written by
Dr. Catherine Shaffer
Catherine Shaffer is a freelance science and health writer from Michigan. She has written for a wide variety of trade and consumer publications on life sciences topics, particularly in the area of drug discovery and development. She holds a Ph.D. in Biological Chemistry and began her career as a laboratory researcher before transitioning to science writing. She also writes and publishes fiction, and in her free time enjoys yoga, biking, and taking care of her pets.
Source: Read Full Article
